비단벌레 (Chrysochroa coreana (fulgidissima))
|
|
▣ 국 명 : 비단벌레 ▣ 학 명 : Chrysochroa coreana Han & Park, 2012 ▣ 지정번호 : 멸종위기 야생생물 Ⅱ급 ▣ 계 통 : 절지동물문 > 곤충강 > 딱정벌레목 > 비단벌레과 |
◎ 형 태 : 몸길이는 3~4 cm 정도이며, 몸 빛깔은 녹색 또는 금색을 띤 녹색이며 광택이 있다. 구릿빛을 띤 자색의 굵은 세로띠가 앞가슴등판과 앞날개에 있다. 몸의 배면은 금색을 띤 녹색이고, 가슴과 배의 중앙부는 금색을 띤 적색이며 매우 화려하다. 암컷과 수컷은 비슷한 형태로 수컷은 겹눈이 튀어 나오고 배 끝이 삼각형으로 패어 있으며, 연한 털이 몸 양쪽에 암컷보다 많이 나 있다.
◎ 생 태 : 비단벌레는 울창한 삼림 지대에 서식한다. 떡갈나무·팽나무·벚나무·느티나무 등 오래된 나뭇가지 끝의 주위를 날아다닌다. 특히 나무 위로 높이 날아다니며 화려한 색깔과 무늬가 빛에 반사되면서 반짝거리는 시각적인 신호로 짝을 찾으며, 짝짓기를 마친 암컷은 애벌레의 먹이가 되는 벚나무, 느티나무, 팽나무 등의 껍질 틈에 알을 낳는다.
◎ 분 포 : 국내에는 전라남북도 해안가 국외에는 일본·중국·대만·인도차이나 반도 등지
① 채집정보
- 채 집 자: 빛고을생태연구소 정헌천
- 채집지역: 전라남도 해남군 삼산면 구림리 산 19-1
◆ 포획성충1 : 느티나무잎에서 휴식 ◆ 포획성충2 : 팽나무잎에서 휴식


◆포획한 성충 2개체

② 채집현황 및 샘플처리
|
비단벌레 채집현황 |
샘플처리 |
|||
|
채집일 |
개체 수 |
DNAseq |
RNAseq |
표본 |
|
2016.08.03 |
2 EA |
1 EA |
1 EA |
- |
③ 기존 NCBI Taxonomy 유전정보 현황
|
유전정보 종류 |
Chrysochroa 속 유전자원 |
Chrysochroa fulgidissima 유전자원 |
|
Nucleotide |
137 |
38 |
|
Protein |
95 |
44 |
|
Genome |
1 |
1 |
|
Gene |
13 |
13 |
④ Sequencing 결과
|
비단벌레 |
|||||
|
Type |
Total Reads |
Total Bases |
Total Bases (Gb) |
GC Rate |
비고 |
|
DNA seq |
325,961,430 |
49,220,175,930 |
49.2 |
37.8 |
|
|
RNA seq |
58,825,814 |
8,882,697,914 |
8.9 |
38.2 |
|
- Illumina DNA Sequncing 결과
|
SOAP de novo |
|||||
|
CC-DNA hiseq |
|||||
|
Input information |
|||||
|
|
|
|
Region1 |
Region2 |
Total |
|
raw data |
|||||
|
Number Of Reads |
|
|
162,980,715 |
162,980,715 |
325,961,430 |
|
Number Of Bases |
|
|
24,610,087,965 |
24,610,087,965 |
49,220,175,930 |
|
error correction |
|||||
|
Number Of Reads |
|
|
162,483,217 |
159,186,968 |
321,670,185 |
|
Number Of Bases |
|
|
24,228,110,547 |
23,100,436,381 |
47,328,546,928 |
|
pair reads |
|||||
|
Number Of Reads |
|
|
|
317,876,854 |
317,876,854 |
|
Number Of Bases |
|
|
|
46,803,883,411 |
46,803,883,411 |
|
single reads |
|||||
|
Number Of Reads |
|
|
|
3,793,331 |
3,793,331 |
|
Number Of Bases |
|
|
|
524,663,517 |
524,663,517 |
|
Assembly Results |
|||||
|
Scaffold Metrics |
> 1 bp* |
> 100 bp |
> 500 bp** |
> 1000 bp |
> 2000 bp |
|
Number Of Scaffolds |
|
1,783,108 |
363,677 |
191,045 |
95,568 |
|
Number Of Bases |
|
971,488,915 |
705,095,978 |
584,974,089 |
451,576,767 |
|
Avg. Scaffold Size |
|
544 |
1,938 |
3,061 |
4,725 |
|
N50 Scaffold Size |
|
1,690 |
3,181 |
4,122 |
5,440 |
|
N80 Scaffold Size |
|
317 |
1,121 |
1,847 |
3,057 |
|
N90 Scaffold Size |
|
157 |
755 |
1,371 |
2,485 |
|
largest Scaffold Size |
|
47,209 |
47,209 |
47,209 |
47,209 |
|
Contig Metrics |
> 1 bp* |
> 100 bp |
> 500 bp** |
> 1000 bp |
> 2000 bp |
|
Number Of Contigs |
36,102,776 |
3,391,809 |
301,263 |
114,746 |
24,916 |
|
Number Of Bases |
2,281,180,119 |
841,230,054 |
324,018,554 |
193,744,414 |
71,970,042 |
|
Avg. Contig Size |
63 |
248 |
1,075 |
1,688 |
2,888 |
|
N50 Contig Size |
67 |
298 |
1,172 |
1,686 |
2,788 |
|
N80 Contig Size |
35 |
134 |
715 |
1,215 |
2,246 |
|
N90 Contig Size |
34 |
114 |
599 |
1,101 |
2,116 |
|
Largest Contig Size |
32,562 |
32,562 |
32,562 |
32,562 |
32,562 |
- Illumina RNA Sequncing 결과
|
Total number of raw reads |
|
|
- Number of sequences |
58,825,814 |
|
- Number of bases |
8,882,697,914 |
|
Contig information |
|
|
- Total number of contig |
82,651 |
|
- Number of bases |
51,199,887 |
|
- Mean length of contig (bp) |
619.4 |
|
- N50 length of contig (bp) |
850 |
|
- GC % of contig |
37.23 |
|
- Largest contig (bp) |
6,779 |
|
- No. of large contigs (≥500bp) |
35,568 |
|
Unigene information |
|
|
- Total number of unigenes |
21,797 |
|
- Number of bases |
20,175,251 |
|
- Mean length of unigene (bp) |
925.5 |
|
- N50 length of unigene (bp) |
1,200 |
|
- GC % of unigene |
37.21 |
|
- Length ranges (bp) |
224 – 8,435 |
2) 현재 상황에 대한 고찰
나) 자생 동물자원 Genome Survey
(1) 고품질 genomic DNA 채취 및 표본확보
- 검수용 DNA는 대부분 만족하였고 멸종위기종인 대상종들은 채집량의 제한으로 인해 확증표본으로 제출가능한 표본을 확보하지 못하여 사진으로 대처하고자 함.
|
검수용 샘플 DNA 농도 및 총 양, 확증표본 현황표 |
|||||
|
Sample 명 |
농도(ng/ul) |
Volume(ul) |
DNA 총량(ug) |
저장장소 |
확증표본 |
|
비단벌레 |
86.629 |
236 |
20.444 |
1XTE pH8.0 |
사진으로대체 |
(2) 기타분석
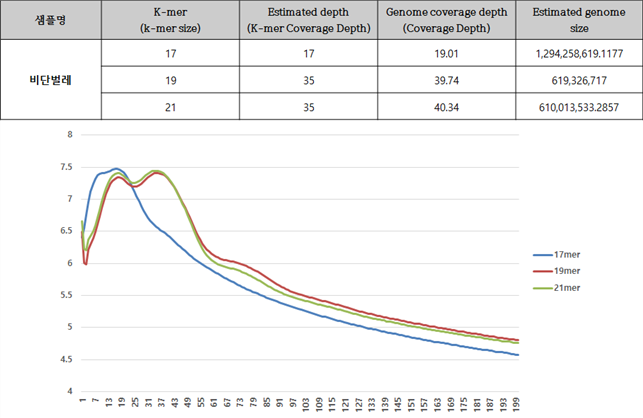
A. Genome Size estimation – Jellyfish 프로그램
각 대상종을 대상으로 Jellyfish 프로그램으로 Genome size를 예측한 결과는 아래와 같다. 각 분석은 k-mer 값 17mer, 19mer, 21mer를 대상으로 하였고 그것을 통해 예측된 genome의 크기는 Estimation genome size 에 표시되었고, 이를 통해 이번 사업에서 분석한 서열의 양의 coverage는 Genome coverage depth(Coverage Depth)에 표기되었다. 그리고 각 종의 표 아래 그래프는 kmer값 별로 나타난 depth의 예측 그래프로 peak값을 확인할 수 있다.
B. 반복서열
- 시퀀싱이 완료된 샘플 DNA 의 contigs 중 1kb 이상의 contigs의 반복서열을 탐색 (Repeat masking results of DNA contigs)
|
비단벌레(Chrysochroa coreana) |
|
비단벌레 (Chrysochroa coreana) 는 1kb 이상의 contig 의 반복서열은 114,746개 read (193,744,414 bp) 이고, GC level 은 36.07% 이다. Repeat Masking 으로 나온 반복서열은 465,929 bp (전체의 0.24%) 만큼 있다. Small RNA 은 607개 read (55,313 bp) 이 있다. |
||||
|
sequences: 114,746 |
|
|||||
|
total length: 193,744,414 bp (193,744,414 bp excl N/X-runs) |
|
|||||
|
GC level : 36.07% |
|
|||||
|
bases masked: 465,929 bp ( 0.24%) |
|
|||||
|
|
number of elements |
length occupied |
percentage of sequence |
|
||
|
SINEs: |
148 |
9,230 bp |
0.00% |
|
||
|
|
ALUs |
0 |
0 bp |
0.00% |
|
|
|
|
MIRs |
6 |
329 bp |
0.00% |
|
|
|
LINEs: |
223 |
18,033 bp |
0.01% |
|
||
|
|
LINE1 |
22 |
1,214 bp |
0.00% |
|
|
|
|
LINE2 |
39 |
2,179 bp |
0.00% |
|
|
|
|
L3/CR1 |
69 |
4,948 bp |
0.00% |
|
|
|
LTRelements: |
46 |
3,047 bp |
0.00% |
|
||
|
|
ERVL |
11 |
654 bp |
0.00% |
|
|
|
|
ERVL-MaLRs |
1 |
57 bp |
0.00% |
|
|
|
|
ERV_classI |
18 |
1,268 bp |
0.00% |
|
|
|
|
ERV_classII |
3 |
207 bp |
0.00% |
|
|
|
DNAelements: |
1,844 |
377,432 bp |
0.19% |
|
||
|
|
hAT-Charlie |
110 |
10,496 bp |
0.01% |
|
|
|
|
TcMar-Tigger |
123 |
16,213 bp |
0.01% |
|
|
|
Unclassified: |
8 |
559 bp |
0.00% |
|
||
|
Total interspersed repeats: |
|
408,301 bp |
0.21% |
|
||
|
Small RNA: |
607 |
55,313 bp |
0.03% |
|
||
|
Satellites: |
36 |
2,365 bp |
0.00% |
|
||
|
|
||||||
C. Mito-genome Map
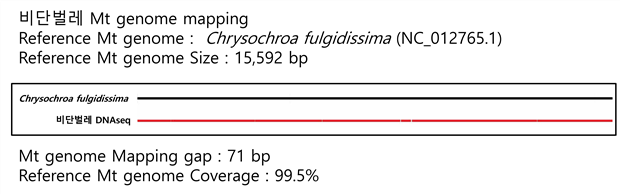
비단벌레는 Reference로 15,592 bp 크기의 Chrysochroa fulgidissima 미토콘드리아 genome을 이용하여 맵핑하였다. 갭은 71 bp 가 나왔고, Coverage 는 99.5%로 확인되었다.
D. Transcriptome data 분석
- 비단벌레
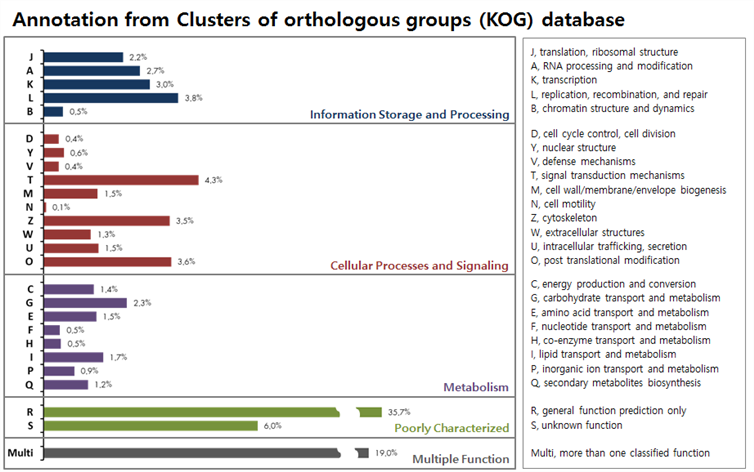
비단벌레의 KOG 결과, “Infomation Storage and Processing” 부분에서 replication/recombination/repair (3.8%) 가 가장 많이 매칭되며, “Cellular Processes and Signaling” 에서는 signal transduction mechanisms (4.3%), post translational modification (3.6%), cytoskeleton (3.5%) 등이 많이 매칭된다. 또한 Metabolism 부분에서 carbohydrate transport and metabolism (2.3 %) 가 가장 많이 매칭되고, Multiple Function 은 19 % 이다.
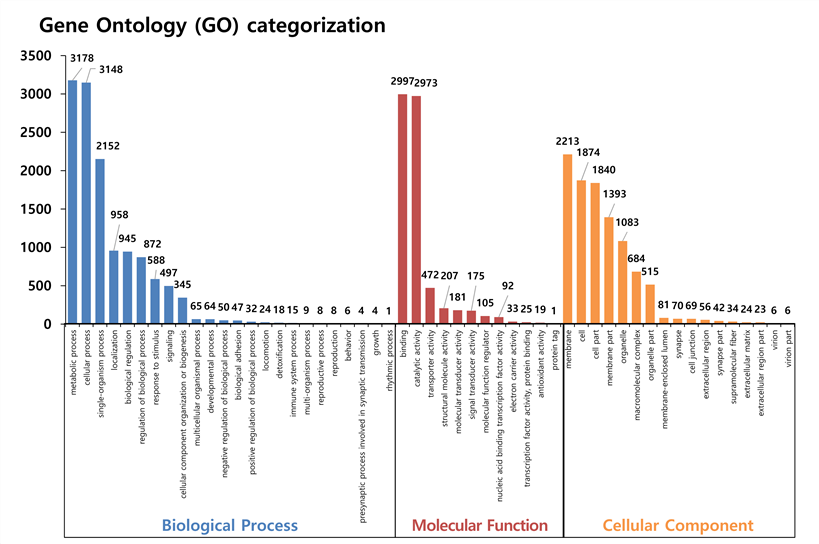
비단벌레의 GO 결과, Biological Process에서 metabolic process (3,178개), cellular process (3,148개) 순으로 많이 매칭된다. Molecular Function 에서는 binding (2,997개), catalytic activity (2,973개) 순으로 가장 많이 매칭된다. 또한, Cellular Component 부분에서는 membrane (2,213개), cell (1,874개), cell part (1,840개) 순으로 많이 매칭된다.
(4) Microsatellite 후보군
A. SSR statistics
|
비단벌레 |
|||||||||||
|
Repeats |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
|
Di |
0 |
0 |
2609 |
2264 |
2050 |
1770 |
1785 |
3152 |
5363 |
1918 |
207 |
|
Tri |
0 |
1686 |
580 |
335 |
294 |
349 |
40 |
2 |
0 |
0 |
0 |
|
Tetra |
1597 |
413 |
148 |
100 |
11 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Penta |
277 |
65 |
22 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Hexa |
37 |
10 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Total |
1911 |
2174 |
3359 |
2699 |
2355 |
2119 |
1825 |
3154 |
5363 |
1918 |
207 |
→ SSR 검색결과 총 27,084개의 SSR 서열을 확인할 수 있었으며, Di motif 가 21,118개로 가장 많이 존재하는 것을 확인 할 수 있었으며, 이를 기반으로 하여 SSR marker 후보군에 대한 primer 를 1,044개 디자인 하였다.
B. Primer sequence
→ 각 종의 유전자를 분석하여 그 종을 구별할 수 있는 primer를 만든다. primer 의 조건은 Motif 약 4~6개이며, Forward 와 Reverse primer 가 18~22개 정도이고 Tm값이 54.5~55.5 정도가 적당하다.
- 비단벌레
|
Sequence |
Motif |
Forward Primer |
Tm |
|
Reverse Primer |
|||
|
71976454 |
(TAAA)6 |
ATGAAATCTTTACACCTCACG |
54.41 |
|
CGAAGTATGGAGGAACAGAAT |
55.91 |
||
|
71977840 |
(ACAA)6 |
CAACCGTTAGACGCTAGAATA |
54.97 |
|
GAGCCCCTTTAATGAAACTAA |
55.33 |
||
|
72009196 |
(AAAT)6 |
ACGGTAGAAAGGCATAAGATT |
54.81 |
|
GCGGACTTCACTAACAATACA |
55.51 |
||
|
72011354 |
(TTTAA)5 |
ATGTGCTGTTGACCAATGTA |
54.87 |
|
CATTCGCATTGAAAGATTCTA |
55.65 |
||
|
72040528 |
(TTTA)4 |
TTTCTTGATTTCAGTTGGTTC |
54.5 |
|
TCTCAATAATCATCGAGGAAG |
54.54 |
||
|
72040862 |
(AATA)5 |
ATCGAGTAACACGCTTAACAG |
54.86 |
|
TATATTTCATGGCATCCTTTG |
55.3 |
||
|
72073202 |
(ATTT)5 |
TACGTAACATTTGGAAAAACC |
54.48 |
|
GTATTGTATTTTTCATTGGAGGT |
54.19 |
||
|
72077006 |
(TGAAC)5 |
TTTATGGCAATTGTGTTTTG |
54.74 |
|
TATTACGGAAGTTTTTGTGGA |
55.02 |
||
|
72099550 |
(TACA)4 |
CAAAGATGGAAGAAATTTGG |
54.87 |
|
AAGCATTTTAGCGGGTATAGT |
55.02 |
||
|
72128528 |
(ACTATT)4 |
ATTGCTTGCACTATTGCTATT |
54.49 |
|
AGCGTATGATAACCGTGAGTA |
55.06 |
||
|
72154646 |
(TTTA)6 |
CGACAAAAAGGTTAAACTGAG |
54.42 |
|
GTGAAACTTTACCAAATGCAC |
54.9 |
||
|
72155614 |
(ACCTA)5 |
CACTCTAAAATTCGCCTACAA |
54.93 |
|
GAGCTTTCATAAAATTCATCG |
54.35 |
||
|
72174704 |
(TTAC)5 |
TGCATGGCGATAGTTTATATT |
55.09 |
|
AGCCGAAACACATGTAGAGTA |
55.09 |
||
|
72195672 |
(AATAT)5 |
CATAAAACATTCCAAGGATCA |
55.17 |
|
TCGTGGGATTGATATGTATGT |
55.31 |
C. 바코드 서열
요번 16년도에 채집한 11종의 유전자를 분석하여 그 종을 구별할 수 있는 Barcode 서열을 만들었고, 그 Barcode 서열을 가지고 NCBI (National Center for Biotechnology Information) 사이트에 있는 BLAST (Basic Local Alignment Search Tool)을 이용하여 결과를 확인하였다. 각 종의 서열들의 BLAST 결과 마커로서 주로 사용되는 COI 서열과 매치되는 것으로 보아 바코드 서열로 사용가능할 것으로 사료된다.
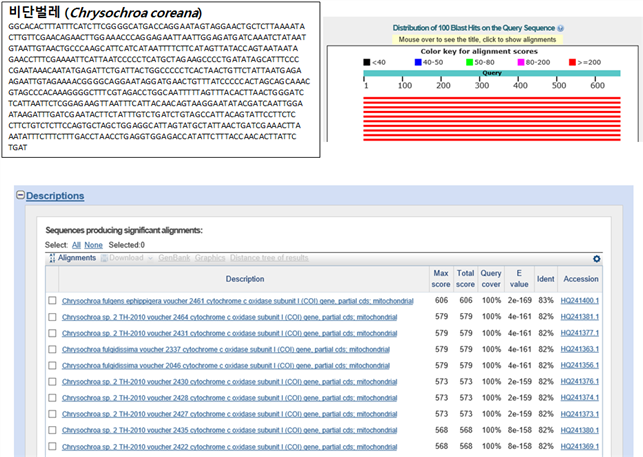
(5) QC 결과
|
DNA Library QC |
|||||
|
Name |
Concentration (ng/ul) |
Volume (ul) |
Quantity (ug) |
Main peak Size (bp) |
Result |
|
비단벌레 |
70.35 |
3.27 |
230.27 |
470 |
pass |
|
RNA Library QC |
|||||
|
Name |
Concentration (ng/ul) |
Volume (ul) |
Quantity (ng) |
Main peak Size (bp) |
Result |
|
비단벌레 |
57.42 |
5.74 |
329.64 |
268 |
Pass |
(6) Gene prediction
|
샘플종 |
Number of Contig (>1kb) |
Number of Bases (>1kb) |
N50 Contig Size |
Number of gene prediction |
|
비단벌레 |
114,746 |
193,744,414 |
1,686 |
56,311 |
각종별로 transcriptome 서열을 대상으로 1kb 이상만을 선택하여 Gene predcition 한 결과 위의 매칭된 contig, base 수는 위의 표와 같고 N50되는 Contig의 길이도 보통 1~3kb 정도로 나타났다. 그리고 이들을 통해 예측된 유전자의 수는 2천개에서 10만개 이상으로 다양하게 예측되었다.
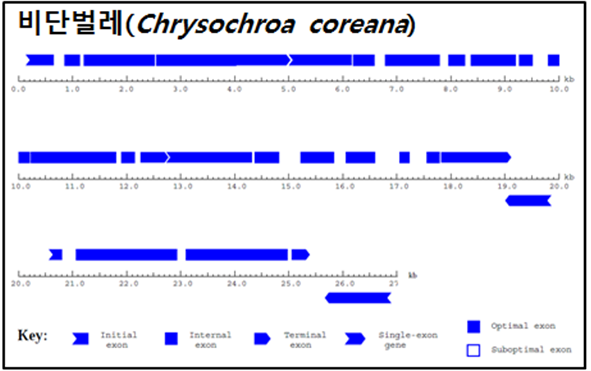
※ Long contig의 Gene prediction 결과 그림 예시