³ªÆÈ°íµÕ (Charonia sauliae)
|
|
¢Ã ±¹ ¸í: ³ªÆÈ°íµÕ ¢Ã ¿µ ¸í: Triton shell ¢Ã ÇÐ ¸í: Charonia sauliae ¢Ã ÁöÁ¤¹øÈ£: ¸êÁ¾À§±âµ¿¹° ¥°±Þ ¢Ã °è Åë: º¹Á·°-Èí°¸ñ-¼ö¿°°íµÕ°ú |
¡Ý »ý±è»õ : Çѱ¹¿¡ »ç´Â º¹Á··ùÁß °¡Àå Å« Á¾À¸·Î ´Ù ÀÚ¶ó¸é ±æÀÌ°¡ 30¿© cm¿¡ ´ÞÇÒ Á¤µµ·Î Ä¿Á® ±¸¸ÛÀ» ¶Õ¾î ³ªÆÈ·Î »ç¿ëÇÏ¿´±â ¶§¹®¿¡ ³ªÆÈ°íµÕÀ̶ó ºÒ¸°´Ù. ¿ø»ÔÇüÀÌ¸ç ³ªÅ¾Àº 7Ãþ¿Ã ³ôÀº ÆíÀÌ´Ù. üÃþ ÁÖº¯¿¡´Â Ȥ ¸ð¾çÀÇ µ¹±â°¡ µÎ ÁÙ ÀÖ´Â °ÍÀÌ Æ¯Â¡ÀÌ´Ù. ³ªÅ¾À» ÀÌ·ç´Â ÃþÁß¿¡¼ À§ÂÊÀÇ ÃþÀº ÁÖȲ»öÀÌ°í, ¾Æ·¡ÂÊ ÃþÀÇ ÁÖ¸§¿¡ ÀÚ°¥»öÀÇ ±¸¸§ ¸ð¾ç ¶Ç´Â Ȳ¹é»ö ¹«´Ì°¡ ±½°Ô ³ª ÀÖ´Ù. ²®µ¥±â ¼ÓÀ¸·Î µé¾î°¡´Â ÁÖµÕÀÌÀÇ ¾ÈÂÊ ¸éÀº Èò»öÀÌ°í, ÁÖµÕÀÌ¿¡¼ ¹Ù±ù³ìÀ¸·Î ³ª¿Í ÀÖ´Â ÀÔ¼úÀº µÎ²¨¿ì¸é¼ ¾à°£ ¹ú¾îÁ® ÀÖ´Ù. ¶Ñ²±Àº µÎ²¨¿î °¢ÁúÀÌ¸ç ¹é»ö ¹ÙÅÁ¿¡ Àû°¥»öÀÇ ¾Æ¸§´Ù¿î ¹Ý¹®À¸·Î µÇ¾î ÀÖÀ¸³ª ÀÔ±¸ ºÎºÐ°ú üÃþÀÇ ÀϺθ¦ Á¦¿ÜÇÏ°í´Â º¸±â Èûµé´Ù.
¡Ý ¼½ÄÁö : Á¶°£´ëºÎÅÍ ¼ö½É ¾à 200m°¡ µÇ´Â ¹Ù´ÙÀÇ ¹ÙÀ§³ª ÀÚ°¥·Î µÈ ¹Ù´Ú¿¡¼ ¼½ÄÇÑ´Ù.
¡Ý ¸ÔÀÌ : À°½Ä¼ºÀ¸·Î ºÒ°¡»ç¸®, ÇØ»ï µîÀÇ ±ØÇǵ¿¹°À» ¸Ô°í »ê´Ù. Çѱ¹¿¡ Á¸ÀçÇÏ´Â ºÒ°¡»ç¸®ÀÇ À¯ÀÏÇÑ ÃµÀûÀ¸·Î ³ªÆÈ°íµÕ ÇÑ ¸¶¸®°¡ ÇÏ·ç¿¡ ºÒ°¡»ç¸® ÇÑ ¸¶¸® ÀÌ»óÀ» ¼·ÃëÇÑ´Ù. À̶§ ÀÌ»¡¿¡ ÇØ´çÇÏ´Â Ä¡¼³À» »ç¿ëÇÏ¸ç ºÒ°¡»ç¸® ÇÑ ¸¶¸®¸¦ Àâ¾Æ¸Ô´Âµ¥ ä 3½Ã°£ÀÌ °É¸®Áö ¾Ê´Â´Ù.
¡Ý ¹ø½Ä : 12¿ù¿¡¼ ´ÙÀ½ÇØ 4¿ù¿¡ °ÉÃÄ »ê¶õÇÑ´Ù.
¡Ý ºÐÆ÷ : ÀϺ», Çʸ®ÇÉ, Çѱ¹ÀÇ ³²Çؾȿ¡ ¼½ÄÇÑ´Ù. ¿¹Àü¿¡´Â Á¦ÁÖµµ, ¿©¼ö µî ÇØ¾È Áö¿ª¿¡¼ ÈçÈ÷ º¼ ¼ö ÀÖ¾úÀ¸³ª ³²È¹ ¹× ¹Ù´Ù·Î Èê·¯µé¾î °£ »ýÈ°Çϼö¿¡ ¿À¿°µÇ¾î °³Ã¼¼ö°¡ ÁÙ¾ú´Ù. ÇöÀç Çѱ¹¿¡¼´Â Á¦ÁÖµµ ±ÙÇØ¿¡¼¸¸ ¼½ÄÇÑ´Ù.
1) äÁýÁ¤º¸
- äÁýÇã°¡: 2014.07.10 / 2¸¶¸®
- ä Áý ÀÚ: °¿ø´ëÇб³ ÀÌÁØ»ó
2) äÁýÇöȲ ¹× »ùÇÃó¸®
|
³ªÆÈ°íµÕ äÁýÇöȲ |
»ùÇÃó¸® |
|||
|
äÁýÀÏ |
°³Ã¼ ¼ö |
DNAseq |
RNAseq |
Ç¥º» |
|
2014.08.28 |
1 EA |
1 EA (¹ß) |
1 EA (³»Àå) |
- |
3) QC Data
|
»ùÇÃÁ¾ |
DNA QC |
RNA QC |
Result |
||
|
Concentration (ug/ul) |
Total Conc. (ug) |
Concentration (ug/ul) |
RIN (>7) |
||
|
³ªÆÈ°íµÕ |
49.3 |
20 |
613 |
6.5 |
pass |
4) ±âÁ¸ NCBI Taxonomy À¯ÀüÁ¤º¸ ÇöȲ
|
À¯ÀüÁ¤º¸ Á¾·ù |
Charonia ¼Ó À¯ÀüÀÚ¿ø |
Charonia sauliae À¯ÀüÀÚ¿ø |
|
Nucleotide |
77 |
7 |
|
Protein |
103 |
40 |
|
Genome |
1 |
1 |
|
Gene |
37 |
37 |
5) Sequencing ºÐ¼® °á°ú
- Illumina DNA Sequncing °á°ú
|
Input information |
|||||
|
¡¡ |
Region1 |
Region2 |
Region3 |
Region4 |
Total |
|
raw data |
|||||
|
Number Of Reads |
93,853,943 |
93,853,943 |
29,317,242 |
29,317,242 |
246,342,370 |
|
Number Of Bases |
11,825,596,818 |
11,825,596,818 |
3,693,972,492 |
3,693,972,492 |
31,039,138,620 |
|
error correction |
|||||
|
Number Of Reads |
78,184,324 |
74,212,243 |
16,787,721 |
16,115,815 |
185,300,103 |
|
Number Of Bases |
7,985,725,569 |
7,392,881,494 |
1,600,734,724 |
1,503,673,595 |
18,483,015,382 |
|
pair reads |
|||||
|
Number Of Reads |
|
130,053,922 |
|
24,029,658 |
154,083,580 |
|
Number Of Bases |
|
13,343,583,209 |
|
2,351,240,874 |
15,694,824,083 |
|
single reads |
|||||
|
Number Of Reads |
|
22,342,645 |
|
8,873,878 |
31,216,523 |
|
Number Of Bases |
|
2,035,023,854 |
|
753,167,445 |
2,788,191,299 |
|
Assembly Results |
|||||
|
Scaffold Metrics |
> 1 bp* |
> 100 bp |
> 500 bp** |
> 1000 bp |
> 2000 bp |
|
Number Of Scaffolds |
1,680,067 |
1,680,067 |
59,333 |
4,974 |
282 |
|
Number Of Bases |
334,439,867 |
334,439,867 |
41,286,796 |
6,569,766 |
727,659 |
|
Avg. Scaffold Size |
199 |
199 |
695 |
1,320 |
2,580 |
|
N50 Scaffold Size |
205 |
205 |
665 |
1,256 |
2,401 |
|
N80 Scaffold Size |
137 |
137 |
550 |
1,076 |
2,123 |
|
N90 Scaffold Size |
114 |
114 |
523 |
1,037 |
2,054 |
|
largest Scaffold Size |
12,993 |
12,993 |
12,993 |
12,993 |
12,993 |
|
Contig Metrics |
> 1 bp* |
> 100 bp |
> 500 bp** |
> 1000 bp |
> 2000 bp |
|
Number Of Contigs |
26,321,282 |
2,141,545 |
11,516 |
969 |
80 |
|
Number Of Bases |
1,694,442,839 |
352,732,303 |
8,011,855 |
1,342,501 |
238,117 |
|
Avg. Contig Size |
64 |
164 |
695 |
1,385 |
2,976 |
|
N50 Contig Size |
57 |
163 |
656 |
1,286 |
2,732 |
|
N80 Contig Size |
47 |
130 |
546 |
1,076 |
2,237 |
|
N90 Contig Size |
46 |
111 |
522 |
1,037 |
2,077 |
|
Largest Contig Size |
12,993 |
12,993 |
12,993 |
12,993 |
12,993 |
- Illumina RNA Sequncing °á°ú
|
Input information |
|||||
|
¡¡ |
¡¡ |
¡¡ |
Region1 |
Region2 |
Total |
|
raw data |
|||||
|
Number Of Reads |
|
|
128,846,778 |
128,846,778 |
257,693,556 |
|
Number Of Bases |
|
|
16,234,694,028 |
16,234,694,028 |
32,469,388,056 |
|
error correction |
|||||
|
Number Of Reads |
|
|
126,149,321 |
125,019,833 |
251,169,154 |
|
Number Of Bases |
|
|
15,419,410,042 |
15,261,273,439 |
30,680,683,481 |
|
pair reads |
|||||
|
Number Of Reads |
|
|
|
|
247,803,298 |
|
Number Of Bases |
|
|
|
|
30,343,009,485 |
|
single reads |
|||||
|
Number Of Reads |
|
|
|
|
3,365,856 |
|
Number Of Bases |
|
|
|
|
337,673,996 |
|
Assembly Results |
|||||
|
Scaffold Metrics |
> 1 bp* |
> 100 bp |
> 500 bp** |
> 1000 bp |
> 2000 bp |
|
Number Of Scaffolds |
334,367 |
334,367 |
60,326 |
23,539 |
7,047 |
|
Number Of Bases |
126,662,755 |
126,662,755 |
70,883,894 |
45,462,610 |
22,845,904 |
|
Avg. Scaffold Size |
378 |
378 |
1,175 |
1,931 |
3,241 |
|
N50 Scaffold Size |
614 |
614 |
1,335 |
2,009 |
3,204 |
|
N80 Scaffold Size |
207 |
207 |
731 |
1,299 |
2,372 |
|
N90 Scaffold Size |
148 |
148 |
606 |
1,140 |
2,177 |
|
largest Scaffold Size |
21,910 |
21,910 |
21,910 |
21,910 |
21,910 |
|
Contig Metrics |
> 1 bp* |
> 100 bp |
> 500 bp** |
> 1000 bp |
> 2000 bp |
|
Number Of Contigs |
4,925,670 |
482,144 |
49,698 |
14,439 |
2,889 |
|
Number Of Bases |
395,122,578 |
132,116,369 |
47,889,673 |
23,980,019 |
8,468,080 |
|
Avg. Contig Size |
80 |
274 |
963 |
1,660 |
2,931 |
|
N50 Contig Size |
79 |
330 |
1,001 |
1,641 |
2,806 |
|
N80 Contig Size |
49 |
160 |
641 |
1,197 |
2,247 |
|
N90 Contig Size |
46 |
132 |
563 |
1,092 |
2,117 |
|
Largest Contig Size |
21,825 |
21,825 |
21,825 |
21,825 |
21,825 |
³ª) ¸êÁ¾À§±âÁ¾ µ¿¹° Genome Survey
|
°Ë¼ö¿ë »ùÇà DNA ³óµµ ¹× ÃÑ ¾ç, È®ÁõÇ¥º» ÇöȲǥ |
|||||||
|
¡¡Sample¸í |
³óµµ(ng/ul) |
Volume(ul) |
DNA ÃÑ·®(ug) |
O.D A260/280 |
È®ÁõÇ¥º» |
äÁýÇã°¡ÀÏ |
äÁýÀÏ |
|
³ªÆÈ°íµÕ |
271 |
996 |
269 |
1.81 |
X |
2014.7.10 |
2014.8.28 |
(2) À¯Àüü ¼¿È®º¸ : ÇöÀç 20Á¾ È®º¸
|
Sample |
±¸ºÐ |
raw data size |
trim data size |
ºñ°í |
|
³ªÆÈ°íµÕ |
DNA |
31,039,138,620 |
18,483,015,382 |
¡¡ |
|
RNA |
32,469,388,056 |
30,680,683,481 |
¡¡ |
(3) ±âŸºÐ¼®
A. Genome Size estimation
|
»ù Çà ¸í |
Kmer size |
Kmer total |
Pk depth |
Genome_size |
Coverage(X) |
|
³ªÆÈ°íµÕ |
23 |
14,641,215,108 |
164 |
89,275,701 |
210.256 |
B. ¹Ýº¹¼¿
- ½ÃÄö½ÌÀÌ ¿Ï·áµÈ ³ªÆÈ°íµÕ DNA ¹× RNAÀÇ contigs Áß 1kb ÀÌ»óÀÇ contigsÀÇ ¹Ýº¹¼¿À» Ž»ö(Left; Repeat masking results of DNA contigs, Right; Repeat masking results of RNA contigs).
|
sequences: 1,061 |
|
sequences: 14,439 |
||||||||
|
total length: 1,457,206 bp (1,457,206 bp excl N/X-runs) |
|
total length: 23,980,019 bp (23,980,019 bp excl N/X-runs) |
||||||||
|
GC level: 39.09% |
|
GC level: 45.32% |
||||||||
|
bases masked: 13,177 bp (0.90%) |
|
bases masked: 34,041 bp (0.14%) |
||||||||
|
|
number of elements |
length occupied |
percentage of sequence |
|
|
number of elements |
length occupied |
percentage of sequence |
||
|
SINEs: |
13 |
667 bp |
0.05% |
|
SINEs: |
113 |
6,823 bp |
0.03% |
||
|
|
ALUs |
0 |
0 bp |
0.00% |
|
|
ALUs |
0 |
0 bp |
0.00% |
|
|
MIRs |
4 |
241 bp |
0.02% |
|
|
MIRs |
25 |
1,574 bp |
0.01% |
|
LINEs: |
35 |
10,661 bp |
0.73% |
|
LINEs: |
87 |
14,947 bp |
0.06% |
||
|
|
LINE1 |
1 |
54 bp |
0.00% |
|
|
LINE1 |
16 |
1,203 bp |
0.01% |
|
|
LINE2 |
4 |
717 bp |
0.05% |
|
|
LINE2 |
13 |
1,055 bp |
0.00% |
|
|
L3/CR1 |
5 |
409 bp |
0.03% |
|
|
L3/CR1 |
19 |
1,424 bp |
0.01% |
|
LTR elements: |
4 |
943 bp |
0.06% |
|
LTR elements: |
17 |
2,102 bp |
0.01% |
||
|
|
ERVL |
0 |
0 bp |
0.00% |
|
|
ERVL |
1 |
33 bp |
0.00% |
|
|
ERVL-MaLRs |
0 |
0 bp |
0.00% |
|
|
ERVL-MaLRs |
0 |
0 bp |
0.00% |
|
|
ERV_classI |
1 |
96 bp |
0.01% |
|
|
ERV_classI |
8 |
602 bp |
0.00% |
|
|
ERV_classII |
0 |
0 bp |
0.00% |
|
|
ERV_classII |
0 |
0 bp |
0.00% |
|
DNA elements: |
7 |
394 bp |
0.03% |
|
DNA elements: |
28 |
1,524 bp |
0.01% |
||
|
|
hAT-Charlie |
0 |
0 bp |
0.00% |
|
|
hAT-Charlie |
1 |
56 bp |
0.00% |
|
|
TcMar-Tigger |
1 |
60 bp |
0.00% |
|
|
TcMar-Tigger |
0 |
0 bp |
0.00% |
|
Unclassified: |
0 |
0 bp |
0.00% |
|
Unclassified: |
1 |
35 bp |
0.00% |
||
|
Total interspersed repeats: |
|
12,665 bp |
0.87% |
|
Total interspersed repeats: |
|
25,431 bp |
0.11% |
||
|
Small RNA: |
9 |
513 bp |
0.04% |
|
Small RNA: |
101 |
8,708 bp |
0.04% |
||
|
Satellites: |
0 |
0 bp |
0.00% |
|
Satellites: |
1 |
86 bp |
0.00% |
||
|
Simple repeats: |
0 |
0 bp |
0.00% |
|
Simple repeats: |
0 |
0 bp |
0.00% |
||
|
Low complexity: |
0 |
0 bp |
0.00% |
|
Low complexity: |
0 |
0 bp |
0.00% |
||
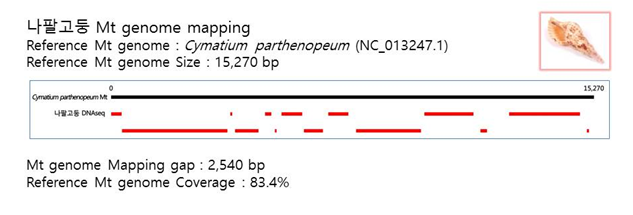
D. Mito-genome Map
(4) Microsatellite È帱º
A. SSR statistics
|
Motif(-mer) |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
Total |
|
³ªÆÈ°íµÕ |
379 |
137 |
73 |
12 |
2 |
- |
- |
- |
- |
603 |
B. ¹ÙÄڵ弿
>³ªÆÈ°íµÕ(Charonia sauliae)
ACTTTATATATTTTATTTGGAATATGAGCGGGGCTAGTAGGAACTGCTCTAAGACTATTA
ATTCGAGCTGAATTAGGTCAACCTGGAGCTTTATTAGGTGACGATCAATTATATAATGTA
ATTGTAACTGCTCATGCTTTTGTAATAATTTTTTTCTTAGTGATACCAATGATAATTGGA
GGATTTGGAAATTGATTAGTACCACTTATATTAGGTGCTCCAGATATAGCATTTCCTCGA
TTAAATAACATAAGATTCTGATTACTGCCACCAGCATTACTTTTATTACTTTCTNCAGCC
GCTGTGGAAAGANNNGTTGGTACCGGATGAACAGTATATCCTCCATTGGCAGGAAACTTA
GCCCATGCTGGAGGTTCGGTTGATTTGGCTATTTTTTCGTTACATCTTGCTGGTGCATCA
TCTATTCTTGGTGCAGTTAATTTTATTACAACCATTATCAATATACGATGGCGAGGAATA
CAATTTGAACGTCTTCCTTTATTTGTATGATCAGTTAAAATTACAGCTGTACTCTTATTA
CTTTCTCTACCAGTTTTAGCTGGTGCTATTACTATATTATTAACGGATCGAAATTTTAAT
ACTGCCTTTTTTGATCCAGCAGGAGGTGGAGACCCTATTTTATATCAACATCT
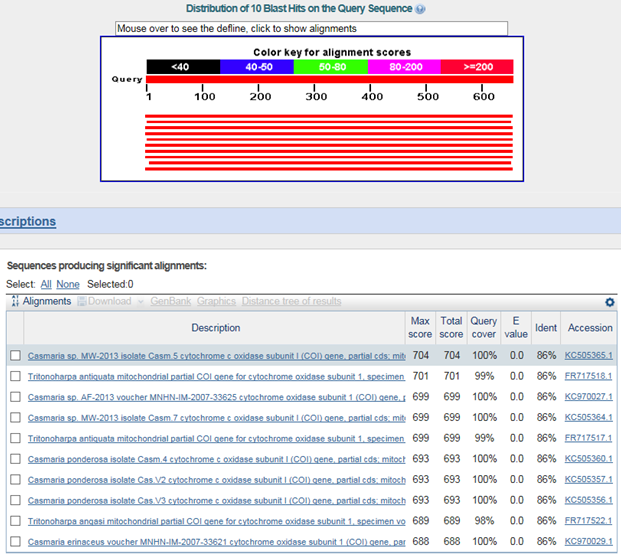
°ËÀºÁ¡ÀÔ¼ú°è¶õ°íµÕ (Casmaria ponderosa) : family ºÎÅÍ ´Ù¸§
Tritonoharpa antiquata : infraorder ºÎÅÍ ´Ù¸§